More Examples
The examples in this section show the properties and operations that may not be as frequently required as others but they show some interesting features of the library.
Circular Nested Data-flow
auto pivot = ezl::flow<char, int>()
.map<2>([](int i) { return i * i; }).colsTransform()
.build();
vector<tuple<char, vector<int>>> buf;
buf.emplace_back(make_tuple('a', vector<int>{2}));
buf.emplace_back(make_tuple('b', vector<int>{3, 4, 5}));
ezl::rise(ezl::fromMem(buf).split())
.map<2>([](const vector<int>& v) {
return ezl::rise(ezl::fromMem(v)).get();
}).colsTransform()
.pipe(pivot) // adds the flow and continues adding to it
.filter<2>(ezl::gt(100)).dump()
.oneUp() // moves to adding to pivot again
.filter<2>(ezl::lt(100))
.pipe(pivot)
.run();We build a pivot dataflow that can process data streams of row type char and int. It takes the second column and transforms the integer to its square.
The buffer has two columns where the second column is a vector.
The data flow loads buffer and applies a map transformation to its second columns. The map returns the result obtained by passing the vector to another dataflow. The dataflow inside the map treats every item in the vector as a row and returns a vector of tuple of integer. Returning a vector implies returning multiple rows. A tuple implies columns. So essentially we are returning multiple rows with a single column. The map has colsTransform property that means that the input column(2nd column) gets replaced by the output column. So for input row (‘b’, vector
Next we add pivot dataflow that we have already built flow to the map unit. After this we add two filters to the map as two branches. We add pivot as the destination of one of the filter. So the rows stream from the pivot to the two filters. If the second column is greater than 100 the first filter let it pass and it gets dumped. If the second column is less than 100 the second filter sends the row back to pivot which then transforms its second column to square and again passes it to the two filters.
Branch and Combine
auto src = rise(fromMem(nums)).build();
auto half = flow<int>().map([](int x) {
return x / 2.;
}).build();
auto twice = flow<int>().map([](int x) {
return x * 2.;
}).build();
auto fl = *(*(src) >> half) + (*(twice) << src);The data stream from source first branches to half and twice and then the output stream of half and twice is merged. Notice, that all the dataflow connections are done using overloaded operators. The same can be achieved using pipe, tee, oneUp and merge dataflow properties.
The ezl::rise takes integer columns from the memory. It streams these numbers to two different branches. The first branch has a map that halves the integers and outputs (number, half). The other branch doubles the integer and outputs (number, twice). The merged flow can then be run or used in any other dataflow.
The pointer deferences with operator overloads look crazy and we are very likely to get rid of them in future versions of easyLambda.
Displaced Atomic Simulation Postprocessor
LAMMPS is a molecular dynamics simulation software. It can deal with very large systems using MPI. While the LAMMPS can scale fairly well on clusters the post-processors written to work with the simulation results may not. The general output from LAMMPS is timestep followed by atomic co-ordinates and other atomic properties like type of atom, id etc for the time-step. ezl fromFile function object has a property lammps which gives each atom with time-step value in every row. It also enables safe parallel reading from a single dump or multiple files.
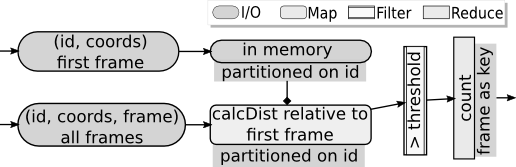
In the following dataflow. We find the number of atoms in every time-step that get displaced more than a threshold value with respect to their position in first frame. The input data-file has atom-id and coordinates in each row and time-step as the header for the rows that follow.
// loading first frame atoms in the memory partitioned on atoms-id.
auto buffer = ezl::rise(ezl::fromFile<int, std::array<float, 3>, int>(argv[1])
.cols({1, 3, 4, 5, 6}) // id, coords
.lammps())
.filter(ezl::tautology()).prll<1>(1.0)
.get();
unordered_map<int, array<float, 3>> firstFrame;
for(const auto& it :buffer) firstFrame[get<0>(it)] = get<1>(it);In the above dataflow we load the atom-ids and coordinates of the atoms in the first frame. We use lammps property to read the rows in the LAMMPS dump. The ezl::tautology function object used in the filter returns true for all the rows. The prll property with partitioning on first column (i.e. id) is used with the filter which runs on all the processes that the rise and in-turn dataflow runs on. The rows are returned to the buffer. Now, the buffer in each process has different set of atom-ids. We then put the atom ids in an unordered_map. Notice that the dataflow works even if the atoms in a single time-step are too big to store on a single system, since the rows are streamed to their respective processes as soon as they are read.
ezl::rise(ezl::fromFile<int, array<float, 3>, int>(argv[2])
.cols({1, 3, 4, 5, 6}) // id, coords, timestep
.lammps())
.map<1, 2>([&firstFrame](int id, array<float, 3> coords) {
return calcDist(coords, firstFrame[id]);
}).prll<1>(1.0).colsTransform()
.filter<1>(ezl::gt(threshold))
.reduce<2>(ezl::count(), 0).hash<hashfn>().inprocess()
.reduce<1>(ezl::sum(), 0).hash<hashfn>()
.reduceAll([](vector<tuple<int, int>> a) {
sort(a.begin(), a.end());
return a;
}).dump(outFile)
.run();
}This dataflow’s rise reads atom-id, coordinates and timesteps for all atoms on all the time-steps. The map function takes as input id and coordinate and returns distance from initial position along with the timestep. The lambda function in the map captures the map created for the co-ordinates of initial time-step in the previous dataflow. The prll property with partitioning on first column is used with the map that runs on all the processes that the rise / dataflow runs on. Since, the paritioning is on id with same default partitioning function, the atom-id that goes to a process in the previous dataflow is certain to go to the same process in this dataflow, as well. Next, the filter makes only the atoms that are displaced more than a certain threshold pass through. Next, we have an inprocess reduce that counts the total number of rows in each time-step. Here, the time-step is the second column. The inprocess reduce outputs rows with (time-step, count). Then, we sum the counts for each time-step (column 1) globally. Notice, that we have a custom hash function for the time-step. This is because generally the time-steps are in multiples of thousand or so and we found that the default hash function does not distribute the time-steps fairly to all the processes. Finally, the reduceAll is used to sort the rows based on time-step for easily reading the output.